Node Overview
Nodes provide template structure for performing data extraction, processing, and API communication. The diagram below shows how a Node performs a base function, but exposes an interface for user-defined code to enable tailoring.
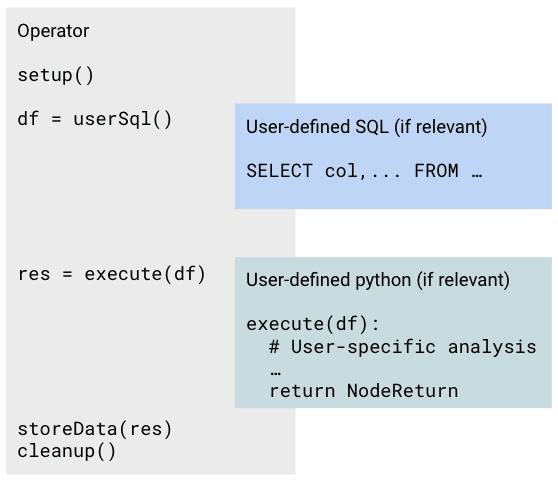
In Jupyter notebooks, Nodes that are editable by users are marked with light blue backgrounds for user-defined SQL and Python cells. These cells can be committed to a repository and deployed to a workflow orchestrator by clicking the button in the toolbar or by pressing Cmd+Shift+S (macOS) or Ctrl+Shift+S (Windows/Linux).
Key Characteristics of Nodes
Understanding how Nodes interact with files, APIs, and their input/output types is essential for effective usage. Nodes may also have a user-editable component, which is indicated in the table below.
List of Available Nodes and Key Characteristics
The table below provides a list of available Nodes, along with details on whether they include a user-editable component and if they can handle multiple inputs.
- Is Editable: Indicates that the Node includes a user-editable component.
- Is Multi: Indicates if the Node can accept multiple inputs.
| Category | Name | Input Types | Output Types | Is Editable | Is Multi |
|---|---|---|---|---|---|
| Analysis | Python | Table(s) and/or File(s) | NodeReturn | True | False |
| Analysis | Transform_SQL | Table(s) | Table | True | False |
| Analysis | Trigger_Python | Table(s) and/or File(s) | FlowInputs | True | False |
| App | API_Node | API | NodeReturn | True | False |
| App | Airtable_Export | Table | API | True | False |
| App | Airtable_Import | API | NodeReturn | True | False |
| App | Azure_Query | Table | NodeReturn | True | False |
| App | Azure_Read | API | FileAny | False | False |
| App | Azure_Write | FileAny | API | False | False |
| App | Benchling_Event | Event | FlowInputs or NodeReturn | True | False |
| App | Benchling_Warehouse_Query | Table | NodeReturn | True | False |
| App | Benchling_Warehouse_Sync | API | NodeReturn | True | False |
| App | Benchling_Write | Table | NodeReturn | True | False |
| App | Benchling_Write_Object | Table(s) and/or File(s) | NodeReturn | True | False |
| App | Coda_Read | Table | NodeReturn | True | False |
| App | Coda_Write | Table | NodeReturn | True | False |
| App | ELabNext_Write | Table | NodeReturn | True | False |
| App | Load_Parquet_to_Table | API | FileAny | True | False |
| App | S3_Event | Event | FlowInputs | True | False |
| App | S3_Read | API | FileAny | True | False |
| App | S3_Write | FileAny | API | False | False |
| App | Smartsheet_Read | API | NodeReturn | True | False |
| App | Snowflake_Write | Table | NodeReturn | False | False |
| App | Webhook_Event | Event | NodeReturn | True | False |
| File | CSV_Read | FileCSV | NodeReturn | True | False |
| File | CSV_Read_Multi | Set[FileCSV] | NodeReturn | True | True |
| File | CSV_Write | Table(s) | NodeReturn | True | False |
| File | Excel_Read | FileExcel | NodeReturn | True | False |
| File | Excel_Read_Multi | Set[FileExcel] | NodeReturn | True | True |
| File | Excel_Write | Table(s) | NodeReturn | True | False |
| File | Image_Read | FileImage | NodeReturn | True | False |
| File | Image_Read_Multi | Set[FileImage] | NodeReturn | True | True |
| File | Image_Write | Table(s) | NodeReturn | True | False |
| File | Input_File | FileAny | NodeReturn | True | False |
| File | Input_File_Multi | Set[FileAny] | NodeReturn | True | True |
| File | PDF_Read | FilePDF | NodeReturn | True | False |
| File | PDF_Read_Multi | Set[FilePDF] | NodeReturn | True | True |
| File | Powerpoint_Write | Table(s) | NodeReturn | True | False |
| File | XML_Read | FileXML | NodeReturn | True | False |
| File | Zip_Read | FileZip | NodeReturn | True | False |
| Instrument | LCMS_Read | File | NodeReturn | True | False |
| Instrument | LCMS_Read_Multi | File | NodeReturn | True | True |
| Instrument | LC_Read | File | NodeReturn | True | False |
| Instrument | LC_Read_Multi | File | NodeReturn | True | True |
| Instrument | WSP_Read | FileWSP | NodeReturn | True | False |
| Tag | Input_Param | string | False | False |
Node Categories
- App: Integrates with third-party APIs for data processing; often requires key exchange between the third-party service and Ganymede.
- Analysis: Performs data manipulations using Python or SQL.
- Instrument: Handles data from laboratory instruments.
- File: Conducts ETL operations on specified data types within the Ganymede cloud.
- Tag: Defines parameters at Flow runtime.
Input and Output Types for Nodes
NodeReturn Object
Many Nodes return a NodeReturn object, which contain tables and files for storage in the Ganymede data lake.
To initialize a NodeReturn object, the following parameters can be passed:
- param tables_to_upload: dict[str, pd.DataFrame] | None - Tables to be stored in Ganymede, keyed by name.
- param files_to_upload: dict[str, bytes] | None - Files to be stored in Ganymede, keyed by filename.
- param if_exists: str - String indicating whether to overwrite or append to existing tables in Ganymede data lake. Valid values are "replace", "append", or "fail"; defaults to "replace".
- param tables_measurement_units: Optional[dict[str, pd.DataFrame]] - (If provided) Specifies the measurement units for columns; keys are table names, values are pandas DataFrames with "column_name" and "unit" as columns.
- param file_location: str - Specifies the bucket location ("input" or "output"); required only if files_to_upload is not null, defaults to "output".
- param wait_for_job: Whether to wait for the write operation to complete before continuing execution; defaults to False.
- param tags: dict[str, list[dict[str, str]] | dict[str, str]] | None: Dictionary of files to tag, with keys as file names and values as a dictionary of keyword parameters for the add_file_tag function. Multiple tags can be added to a single file by passing a list of add_file_tag parameters in the dictionary.
NodeReturn Example
The contents of a NodeReturn object can be inspected in the notebook, where table headers and list of files are displayed. Below is an example of creating a NodeReturn object:
import pandas as pd
def execute():
message = "Message to store in file"
byte_message = bytes(message, "utf-8")
df = pd.DataFrame({"col1": [1, 2, 3], "col2": [4, 5, 6]})
# upload a table named 'my_table' and a file named 'my_file'
return NodeReturn(files_to_upload={"my_file.txt": message}, tables_to_upload={"my_table": df})
execute()
This code produces the following summary of the NodeReturn object:

Docstrings and source code can be viewed by typing ?NodeReturn and ??NodeReturn respectively in a cell in the editor notebook.
Example: NodeReturn Object with Tags
The following code demonstrates the use of the tags parameter in a NodeReturn object:
import pandas as pd
def execute():
message = "Message to store in file"
byte_message = bytes(message, "utf-8")
df = pd.DataFrame({"col1": [1, 2, 3], "col2": [4, 5, 6]})
# Tags are added to the file 'my_file.txt'
# Any parameters that can be passed into the add_file_tag function can be passed into the tags parameter
# of the NodeReturn object. For more information on the add_file_tag function, see the Tags page.
#
# Note that the input_file_path parameter within the add_file_tag function does not need to be specified
return NodeReturn(files_to_upload={"my_file.txt": message}, tables_to_upload={"my_table": df},
tags={"my_file.txt": [{"tag_type_id": "Experiment ID", "display_value": "EXP005"}]})
execute()
FlowInputs object
Nodes that trigger other Flows return a FlowInputs object, which specifies the inputs to the triggered Flow.
To initialize a FlowInputs object, use the following parameters from ganymede_sdk.io:
- param files: list[FlowInputFile] | None - Files to pass to the triggered Flow.
- param params: list[FlowInputParam] | None - Parameters to pass to triggered Flow.
- param tags: list[Tag] | None - Tags to pass to the triggered Flow.
FlowInputFile is a dataclass used for passing files to a Node. Attributes include:
- param node_name: str - Name of the Node within triggered Flow to pass file(s) into
- param param_name: str - Node parameter in the triggered Flow Node that specifies the string pattern that the filename must match (e.g., "csv" for the CSV_Read Node)
- param files: dict[str, bytes] - Files to pass into Node
FlowInputParam is a dataclass used to pass parameters into a Node. It has the following attributes:
- param node_name: str - Name of the Node within triggered Flow to pass parameter(s) to.
- param param_name: str - Node parameter in the triggered Flow Node that is used to specify the string pattern that the parameter must match (e.g., "param" for the Input_Param Node).
- param param_value: str - Value to pass into Node.
Other input/output types
Some other input and output types characteristic to Nodes are:
- Table: Tabular data retrieved from or passed to the Ganymede data lake via ANSI SQL queries or as Pandas DataFrames.
- API: Access through third-party APIs.
- File-related inputs/outputs: Specific file types, including:
- FileAVI: AVI file
- FileCSV: CSV file
- FileExcel: Excel file (xls, xlsx, ..)
- FileImage: Image file (png, bmp, ..)
- FileXML: XML file
- FileZip: Zip file
- FileAny: generic data file, which may be unstructured
- string: String parameter
Python sets, lists, and dictionaries are denoted as Set, List, and Dict, respectively.
Optional indicates that the input or output is optional.
User-editable Nodes
User-editable Nodes present an interface for modifying and testing code that is executed by the workflow management system. These Jupyter notebooks are split into the following sections:
- Node Description: A brief description of the Node's functionality.
- Node Input Data: For Nodes that retrieve tabular data, this section specifies the SQL query used to fetch data for processing.
- User-Defined Function: The execute function in this section processes the data. The function is called during the Flow execution.
The execute function may call classes and functions found within the User-Defined Function cell.
- Testing Section: Cells in this section are for testing modifications to the SQL query and user-defined Python function. After making edits, save changes by clicking the button in the toolbar or selecting "Save Commit and Deploy" from the Kernel menu.
List of Available Nodes
| Category | Name | Brief Description |
|---|---|---|
| Analysis | Python | Process data with python |
| Analysis | Transform_SQL | SQL analysis Function |
| Analysis | Trigger_Python | Process data with Python and trigger subsequent flow |
| App | API_Node | Generic API Access Node |
| App | Airtable_Export | Export data from Ganymede data lake to Airtable |
| App | Airtable_Import | Import data from Airtable into Ganymede data lake |
| App | Azure_Query | Query data from Azure SQL Server |
| App | Azure_Read | Read data from Azure Blob Storage |
| App | Azure_Write | Write data to Azure Blob storage |
| App | Benchling_Event | Capture events from Benchling for triggering flows or saving data |
| App | Benchling_Warehouse_Query | Query Benchling Warehouse from Ganymede |
| App | Benchling_Warehouse_Sync | Sync Benchling Warehouse to Ganymede |
| App | Benchling_Write | Write to Benchling |
| App | Benchling_Write_Object | Write object to Benchling |
| App | Coda_Read | Read Coda tables |
| App | Coda_Write | Write Coda tables |
| App | ELabNext_Write | Create and write eLabNext entry |
| App | Load_Parquet_to_Table | Create data lake table from parquet files |
| App | S3_Event | Capture events from AWS S3 for triggering flows |
| App | S3_Read | Ingest data into Ganymede data storage from AWS S3 storage |
| App | S3_Write | Write data to an S3 bucket |
| App | Smartsheet_Read | Read sheet from Smartsheet |
| App | Snowflake_Write | Sync tables in Ganymede data lake to Snowflake |
| App | Webhook_Event | Capture events from a webhook for triggering flows |
| File | CSV_Read | Read in contents of a CSV file |
| File | CSV_Read_Multi | Read in contents of multiple CSV files |
| File | CSV_Write | Write table to CSV file |
| File | Excel_Read | Read Excel spreadsheet |
| File | Excel_Read_Multi | Read Excel spreadsheets |
| File | Excel_Write | Write Excel spreadsheet |
| File | Image_Read | Process image data; store processed images to data store |
| File | Image_Read_Multi | Process image data for multiple images; store processed images to data store |
| File | Image_Write | Process tabular data; write an image to data lake |
| File | Input_File | Read data file and process in Ganymede |
| File | Input_File_Multi | Read data files and process in Ganymede |
| File | PDF_Read | Read in contents of an PDF file to a table |
| File | PDF_Read_Multi | Read in contents of multiple pdf files to a table |
| File | Powerpoint_Write | Process tabular data; write a powerpoint presentation to data lake |
| File | XML_Read | Read XML file into data lake |
| File | Zip_Read | Extract Zip file |
| Instrument | LCMS_Read | Read and process LCMS file in mzML format |
| Instrument | LCMS_Read_Multi | Read and process multiple LCMS files |
| Instrument | LC_Read | Read and process an Agilent Chemstation / MassStation HPLC file |
| Instrument | LC_Read_Multi | Read and process multiple Agilent Chemstation / MassStation HPLC files |
| Instrument | WSP_Read | Read FlowJo WSP file into data lake |
| Tag | Input_Param | Input parameter into Flow |