Node Editor
Node Overview
Nodes are customizable blocks of code that execute specific tasks within a Flow. Each node in Ganymede provides a template to define and test pipeline logic.
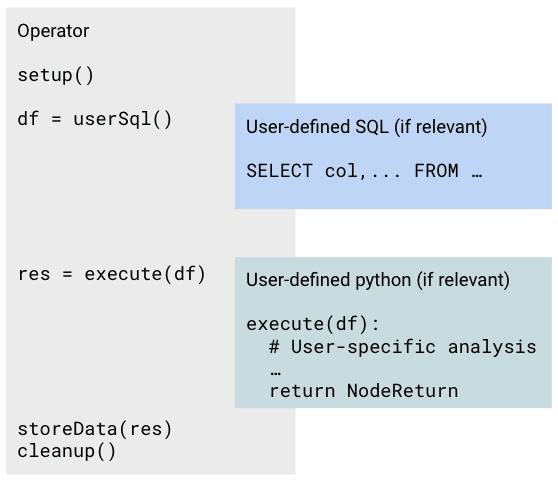
On the Flow Editor page, nodes are displayed as boxes with headers containing the node name. You can edit the node name by clicking on the header.
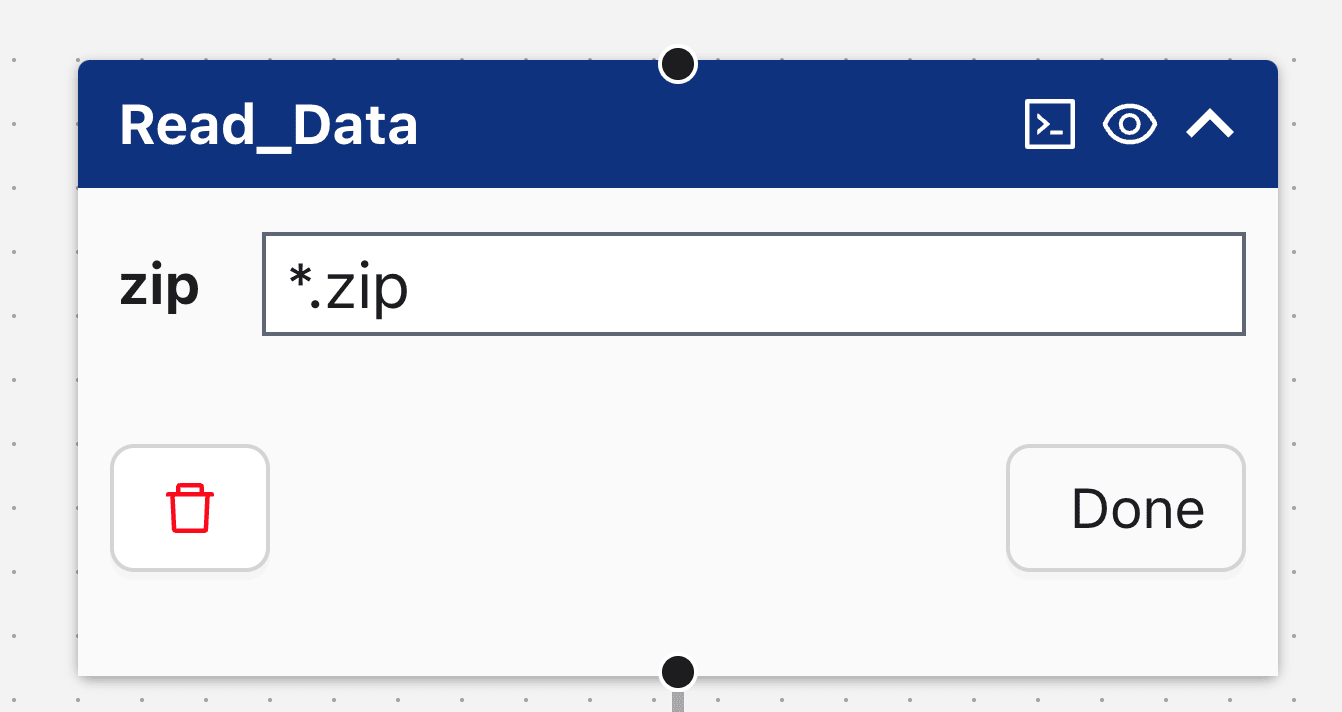
Each node in the Flow Editor includes:
- Node Name: Located in the header; in the example above, it's "CSV_Read."
- Terminal Icon: if present, the is a button that opens the user-editable notebook when clicked.
- Node Attribute(s): If present, these attributes are processed by the node. In the image above, the node attributes "csv" and "results" have values ".csv" and "example_results," respectively.
By default, nodes are shown in a collapsed state. Click the chevron in the upper-right corner to expand the node and view its attributes.
Nodes with editable components open a notebook with a common structure:
Cells Saved on Notebook Save
- User-defined SQL: Retrieves tabular data to be processed by the node. This code is executed by the workflow management system.
- User-defined Python: Processes input data. This code is also executed by the workflow management system.
These cells have a light blue background.
Cells Discarded Upon Notebook Close
- Testing Section: Allows you to test changes to the user-defined SQL and Python before committing and deploying the code.
Cells in this section have a light gray background.
Cells created in the notebook have a light gray background, and are discarded when the webpage for the notebook is closed.
A full listing of available nodes and their key characteristics can be found on the Node Overview page.
Backing Notebooks
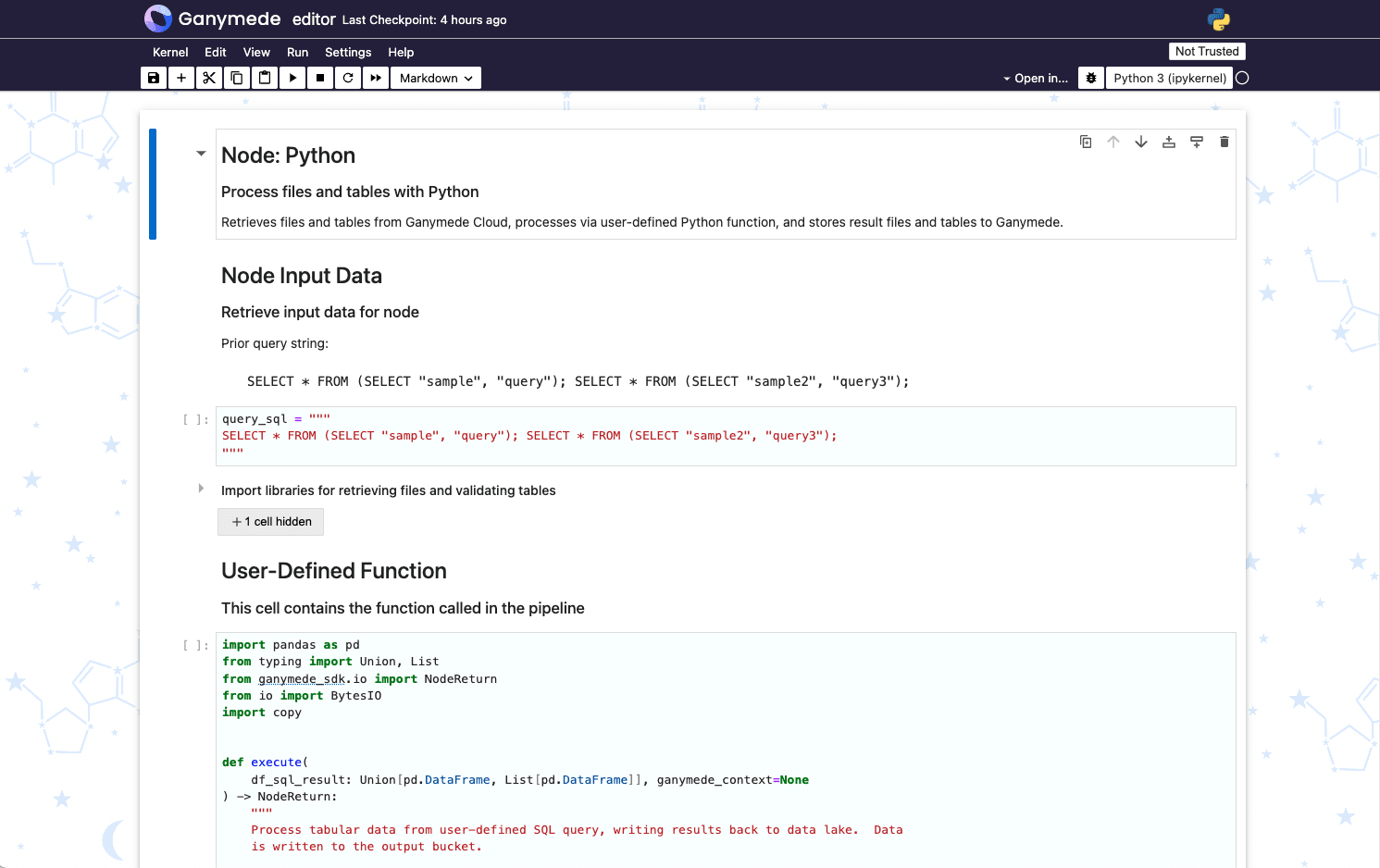
Editable nodes contain a terminal icon () in their upper-right corner. Clicking this icon opens the notebook backing the node. These notebooks format code upon cell execution using black, and perform type checking, hinting, and code diagnostics using Jupyter LSP configured with Ruff.
Code Warnings and Errors
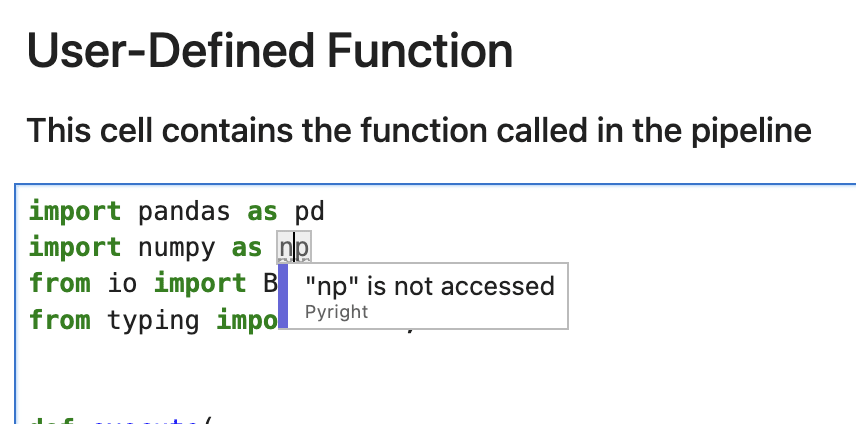
Hover over any underlined piece of code to see warnings and errors from the LSP:
Viewing Class or Method Signatures
Hover over a class or method and click Ctrl to display a tooltip with the class or method signature:
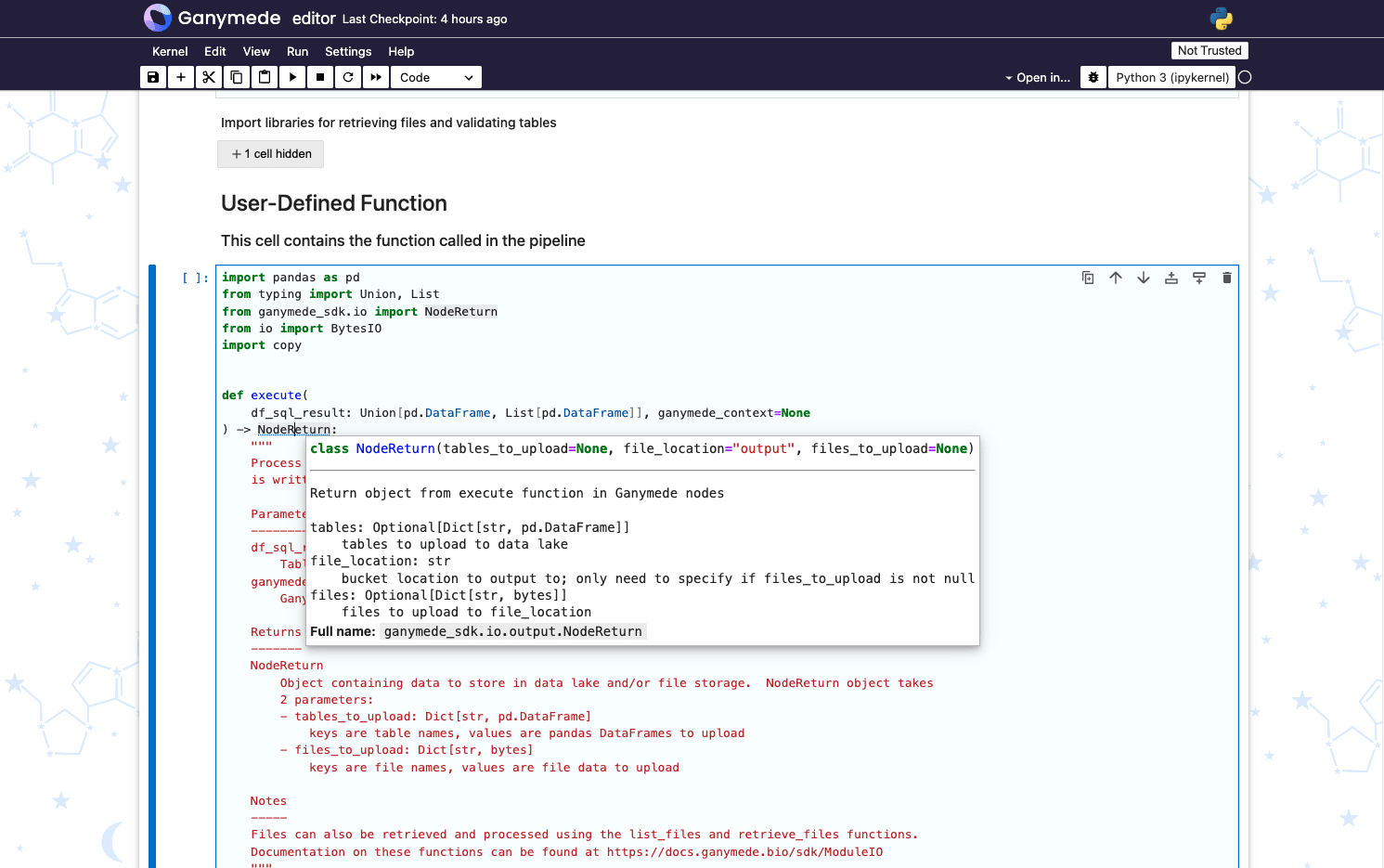
Pipeline Code Notebook Sections
User-defined SQL
query_sql = """
SELECT * FROM demo_file;
"""
For nodes that reference the Ganymede data lake on input, the query_sql string is used to specify the data table to process on the input parameter in the execute function. The schema in the query is specific to the tenant used, and the file corresponds to the name used in an upstream node.
Available tables and their associated schemas can be observed in the Data Explorer tab of the Ganymede app or explored in Dashboards.
Multiple queries can be specified as input by separating them with semicolons. These queries are passed as a list of tables to the user-defined Python function.
User-defined Python
import pandas as pd
from typing import Union, List
from io import BytesIO
from ganymede_sdk.io import NodeReturn
def execute(
df_sql_result: Union[pd.DataFrame, List[pd.DataFrame]], ganymede_context=None
) -> NodeReturn:
"""
Process tabular data from user-defined SQL query, writing results back to data lake. Data
is written to the output bucket.
Parameters
----------
df_sql_result : Union[pd.DataFrame, List[pd.DataFrame]]
Table(s) or list of tables retrieved from user-defined SQL query
ganymede_context : GanymedeContext
Ganymede context variable, which stores flow run metadata
Returns
-------
NodeReturn
Object containing data to store in data lake and/or file storage. NodeReturn object takes
2 parameters:
- tables_to_upload: Dict[str, pd.DataFrame]
keys are table names, values are pandas DataFrames to upload
- files_to_upload: Dict[str, bytes]
keys are file names, values are file data to upload
Notes
-----
Files can also be retrieved and processed using the list_files and retrieve_files functions.
Documentation on these functions can be found at https://docs.ganymede.bio/sdk/ModuleIO
"""
# Retrieve single table from SQL query into a dictionary
if isinstance(df_sql_result, pd.DataFrame):
dict_df_out = {"results": df_sql_result.copy()}
table_out = df_sql_result
else:
dict_df_out = {str(k): df for k, df in enumerate(df_sql_result)}
table_out = df_sql_result[0]
# Writes contents of table to CSV file
bio = BytesIO()
table_out.to_csv(bio)
bio.seek(0)
files_out = {"demo_file": bio.read()}
# Return object containing data to store in data lake and/or file storage
# files_to_upload is a dictionary of file names and file data to upload
# tables_to_upload is a dictionary of table names and pandas DataFrames to upload
#
# For more information on the NodeReturn object, type ?NodeReturn or ??NodeReturn in a cell
return NodeReturn(
files_to_upload=files_out,
tables_to_upload=dict_df_out,
)
In this example, the execute function takes the results of 1 or more tables as input, presented as either a pandas DataFrame or list of pandas DataFrames.
A single table is retrieved from df_sql_result, and the results are written to both the tabular database and a CSV file.
The names of the query_sql variable and execute function cannot be modified or moved to different cells in a notebook. When saving the notebook, these cells are saved, and all other cells are discarded.
This allows users to test in other cells before committing and deploying the code.
Testing Section
# Example 1: Testing section for Python node
from ganymede_sdk import Ganymede
# Declare Ganymede object, which retrieves Ganymede context object from prior run to enable mimicking the execution environment of the most recent pipeline run
g = Ganymede()
# Retrieve dataframes to process; the context object enables rendering of Jinja template variables used in data retrieval
# More detail can be found on the Flow Context & Metadata page of this documentation, under the Template Variables section
result_dfs = g.retrieve_sql(query_sql)
result_dfs
# Run user-defined python on retrieved data
res = execute(result_dfs, ganymede_context=g.ganymede_context)
res
The testing section enables users to test pipeline code prior to deployment. In the example above, the testing code executes query_sql and sends the result to the execute function.
Nodes that load data use a file upload widget so that a file can be uploaded prior to processing. Running the code below yields a button that allows users to submit a CSV file. Data associated with the code can be accessed by the execute function as shown in the 2nd code snippet below.
# Example 2: Testing section for Input_File node
# Create widget for uploading CSV for testing node operation
from IPython.display import display
import ipywidgets as widgets
# Create file upload widget
file_uploader = widgets.FileUpload(
multiple=False,
)
display(file_uploader)
# ---
# Retrieve context variable from most recent run
from ganymede_sdk import Ganymede
g = Ganymede()
# Assemble file uploaded into dictionary keyed by filename
f = file_uploader.value[0]
input_file = {f.name: f.content.tobytes()}
print("\nInput file: \n" + "\n".join(input_file.keys()))
# ---
# Run user-defined Python on uploaded file
res = execute(input_file, ganymede_context=g.ganymede_context)
res
Saving and Deploying Pipeline Code
You can save notebooks in a few ways:
- Clicking the button in the upper right hand corner of the notebook.
- Typing
Cmd+Shift+S(Mac) orCtrl+Shift+S(Windows/Linux) to save the pipeline cell(s). - Selecting the Kernel menu and then selecting Save Commit and Deploy.
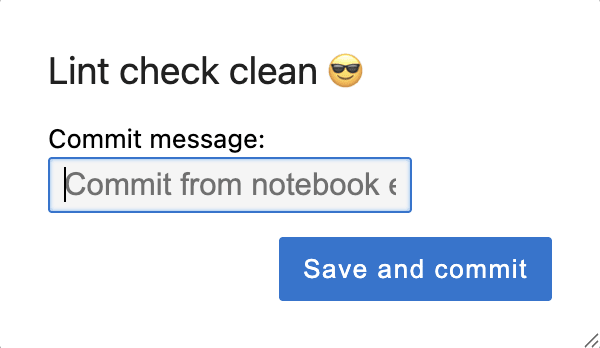
Executing any of these actions will lint the pipeline cell(s). If the code passes linting, the following pop-up will be shown, allowing you to customize a commit message, and pipeline code cells will be saved and deployed.
If the code does not pass linting, the following pop-up will be shown. You can click on the Cancel button to return to the notebook and correct the errors, or Save and Commit to commit the code.
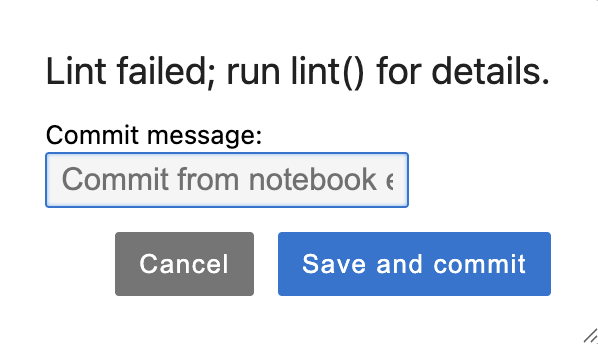
Code is committed and deployed upon saving, even if it does not pass linting, and even if it is not executed prior to saving. This allows you to step outside the structure enforced by black and ruff if desired.
Adding Flow inputs
Flows can be configured to accept user inputs at the start of each run by selecting the appropriate node and saving the Flow. There are three types of inputs:
- File inputs (e.g. - FileAny, FileCSV, etc.)
- Text (string) inputs (e.g. - string)
- Tag inputs (e.g. - Input_Param)
A list of available nodes and their associated categories can be found on the table listing of node characteristics More detail can be found on the Node documentation page.