Tagging Files
What are File Tags?
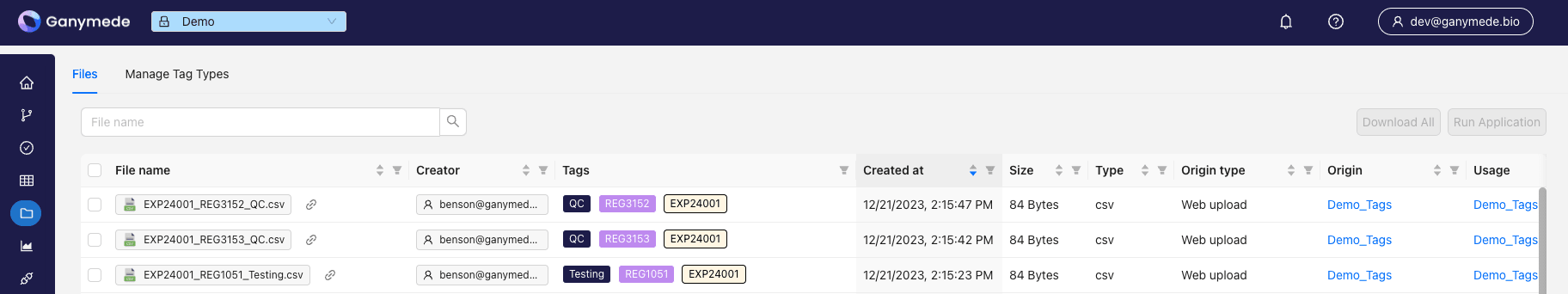
File tags in Ganymede provide a way to organize and categorize files systematically. These tags, specified in user-defined code, allow for efficient filtering and searching within the file browser. Tags can be used to capture various details about a file, such as sample ID, run purpose, experiment ID, experiment step, and any other relevant characteristic defined by users in Ganymede.
Once a file is tagged, it can be easily filtered in the file browser, enabling streamlined access to specific data.
Configuring File Tag Types
Before tagging files, a file tag type must be configured. This is done in the Manage Tag Types section on the Files page.
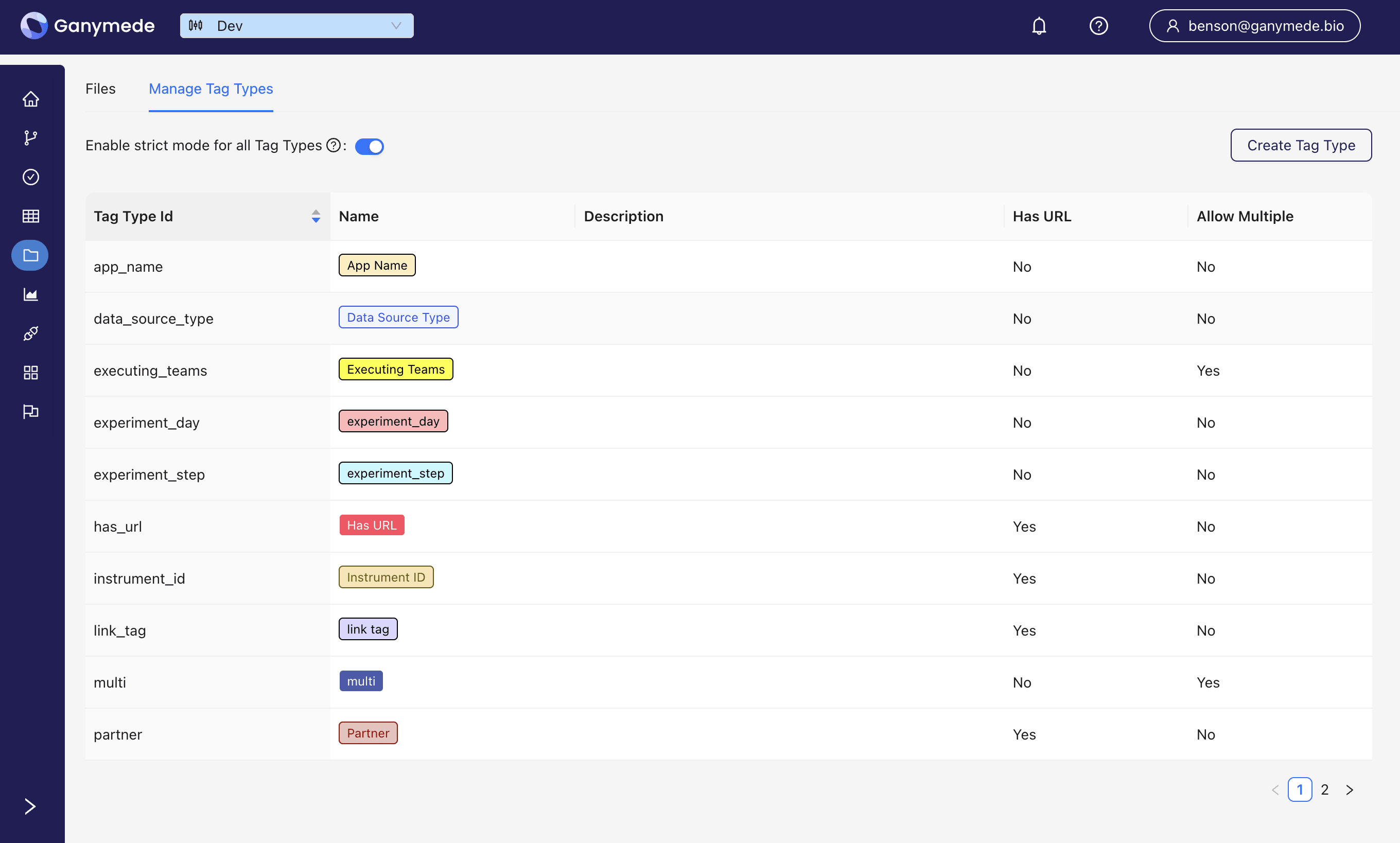
To create a new tag type, click the
button in the upper right corner of the page. This opens a modal where you can specify the attributes of the tag type.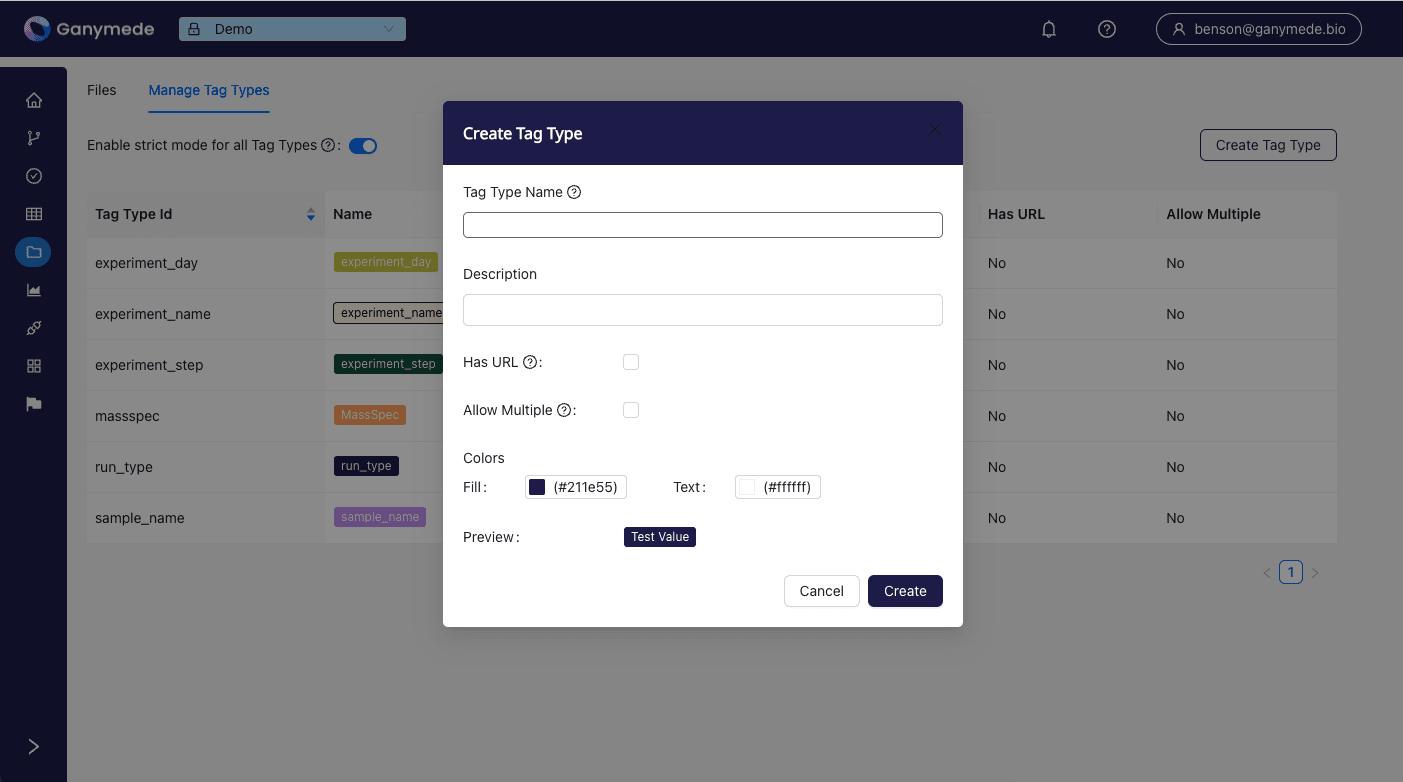
For each tag type, the following attributes can be specified:
- Tag Type Name: The name of the tag type.
- Description: A brief description of the tag type.
- Has URL: Indicates whether the tag value must be a URL (rendered as a link in the file browser).
- Allow Multiple: Allows multiple tags of this type to be applied to a single file.
- Fill: The background color for tags of this type.
- Text: The text color for tags of this type.
The strict mode setting, if disabled, allows admins to delete or modify tags. Tags may only be deleted if they are not applied to any files; the delete button will be grayed out if tag type deletion is not permitted.
Tagging Files
Files can be tagged in user-defined code within Flows and Agents, though the methods differ slightly. In Flows, files are tagged by passing the file path to the add_file_tag function. Within Agents, files are tagged either in the Connections UI, or by passing the FileParam object into the add_file_tag_to_fileparam function. The FileParam object contains the file that the Agent submits to Ganymede storage (for initiating a flow if the Agent is configured to do so).
The full set of methods available for programmatically interacting with tags can be found on the File Tag module in the SDK documentation.
Tagging files in Flow code
To tag a file within a flow, use the add_file_tag function in the ganymede_sdk.file_tag module. This function takes the following arguments:
- param input_file_path: str - The name of the file to tag. The file path can be obtained from the keys in the dictionary returned by the retrieve_files for Ganymede class or by calling the get_gcs_uri method.
- param tag_type_id: str - The name of the tag type to apply to the file.
- param display_value: str - Value of the tag to display in the file browser.
- param tag_id: Optional[str] - The tag id that acts as a unique identifier for the tag.
- param url: Optional[str] - URL to associate with the tag, if applicable.
- param bucket: Optional[str] - The bucket to tag the file in; either "input" or "output". If not specified, the full path to the file in storage must be provided.
from typing import Dict
from ganymede_sdk.file_tag import add_file_tag
from ganymede_sdk.io import NodeReturn
def execute(file_data: Dict[str, bytes], ganymede_context) -> NodeReturn:
entry_name = "test_entry"
for filename in file_data.keys():
add_file_tag(
input_file_path=filename,
tag_type_id="benchling_entry_id",
display_value=entry_name,
bucket="input",
)
File tags can also be added to files returned in the NodeReturn object:
from typing import Dict
from ganymede_sdk.file_tag import add_file_tag
from ganymede_sdk.io import NodeReturn
def execute(file_data: Dict[str, bytes], ganymede_context) -> NodeReturn:
entry_name = "test_entry"
file_data_bytes = list(file_data.values())[0]
return NodeReturn(
files_to_upload={"output_filename.csv": file_data_bytes},
# Parameters for the add_file_tag function are passed in the tags parameter
#
# Note that the input_file_path parameter within the add_file_tag function does not need to be specified
tags={
"output_filename.csv": {
"tag_type_id": "benchling_entry_id",
"display_value": entry_name
}
}
)
Tagging files captured by Agents
From the Connections UI
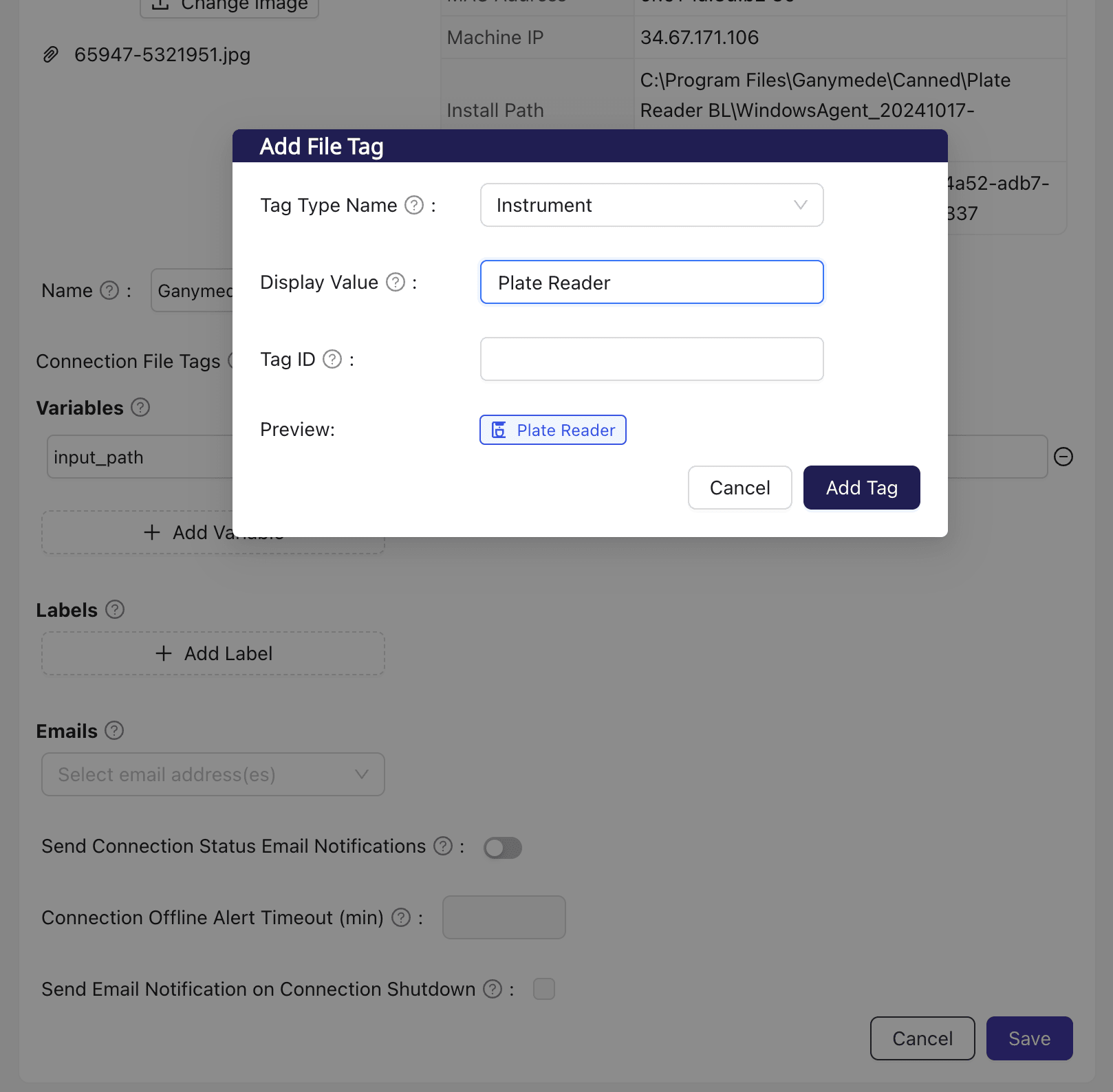
To configure file tagging from the Connections UI, navigate to the relevant Connection on the Connections page. In the Connection Settings box, click on
, followed by .Doing so opens up the modal shown below, where you can specify the tag type and value to apply to files delivered by the Connection.
From Agent user-defined code
To tag a file delivered by an Agent to Ganymede, you can configure an Agent or Connection to tag any files submitted from it using the File tags parameter in Agent configuration.
For example, you might configure the agent to automatically tag files with an instrument ID and a connection from that agent to tag files with a lab location.
You can also tag files programmatically using the add_file_tag_to_fileparam function from the ganymede_sdk.agent module. This function takes the following arguments:
- param file_param: FileParam | MultiFileParam - The FileParam object to tag. FileParam objects hold a single file, while MultiFileParam objects hold multiple files (for use in nodes that take in multiple files). If a MultiFileParam object is specified, the tag is applied to all files within the object.
- param tag_type_id: str - The name of the tag type to apply to the file.
- param display_value: str - The value of the tag to display in the file browser.
- param tag_id: Optional[str] - The tag id that acts as a unique identifier for the tag.
- param url: str - A URL to associate with the tag, if applicable.
from agent_sdk import (
FileParam,
FileWatcherResult,
MultiFileParam,
TriggerFlowParams,
add_file_tag_to_fileparam,
)
# pre v5.0
# from ganymede_sdk.agent import (
# FileParam,
# FileWatcherResult,
# MultiFileParam,
# TriggerFlowParams,
# add_file_tag_to_fileparam,
# )
def execute(flow_params_fw: FileWatcherResult, **kwargs) -> TriggerFlowParams:
"""
Called when all glob patterns specified by get_param_mapping have been matched.
Parameters
----------
flow_params_fw : FileWatcherResult
Dict of FileParam objects indexed by <node name>.<param name>
"""
single_file_params = {}
multi_file_params = {}
labels = kwargs.get("labels", [])
var = kwargs.get("vars", []).get("input_path", "")
for param, files in flow_params_fw.files.items():
if isinstance(files, FileParam):
tagged_file_param = add_file_tag_to_fileparam(files, "lab-agent", var)
for label in labels:
tagged_file_param = add_file_tag_to_fileparam(tagged_file_param, "lab-agent" , label)
single_file_params[param] = tagged_file_param
else:
multi_file_params[param] = MultiFileParam.from_file_param(files)
Files captured by Agents can be tagged via either the Connections page or by code, but should not be tagged in both spots.
Files are passed to and from Virtualization environments using Agents. Therefore, files passed to and from Virtualization environments can be tagged as well.
Note that Virtualization stores a json file in the C:/Program Files/Ganymede/ directory that contains information about the session, including the input files and the current session id.
import json
from pathlib import Path
from agent_sdk import FileWatcherResult, NoOpFileTagParams, add_file_tag_to_fileparam
from ganymede_sdk.file_tag import get_file_tags
SESSION_JSON_PATH = Path("C:/Program Files/Ganymede/session.json")
# Virtualization agent example for that tagging all files sent to Ganymede cloud storage with a notebook entry ID.
def execute(flow_params_fw: FileWatcherResult, **kwargs) -> NoOpFileTagParams:
file_params = list(flow_params_fw.files.values())
with open(SESSION_JSON_PATH, "r") as f:
session_json = f.read()
session_dict = json.loads(session_json)
input_files = session_dict["input_files"]
entry_tags = []
# A tag type defined in this tenant
NOTEBOOK_ENTRY_TAG_TYPE_ID = "notebook_entry_id"
for file in input_files:
tags = get_file_tags(file)
if tags is None:
continue
for tag in tags:
if tag.tag_type_id == NOTEBOOK_ENTRY_TAG_TYPE_ID:
entry_tags.append(tag)
if len(entry_tags):
entry_tag = entry_tags[0]
for file_param in file_params:
add_file_tag_to_fileparam(
file_param=file_param,
tag_type_id=NOTEBOOK_ENTRY_TAG_TYPE_ID,
display_value=entry_tag.display_value,
url=entry_tag.url,
)
return NoOpFileTagParams(files=file_params)